


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية| Telomeres Seal the Chromosome Ends and Function in Meiotic Chromosome Pairing |
|
|
|
Read More
Date: 17-4-2021
Date: 29-5-2021
Date: 28-4-2016
|
Telomeres Seal the Chromosome Ends and Function in Meiotic Chromosome Pairing
KEY CONCEPTS
- The protein TRF2 catalyzes a reaction in which the 3′ repeating unit of the G+T–rich strand forms a loop by displacing its homolog in an upstream region of the telomere.
- Telomeres promote pairing, synapsis, and recombination during meiosis via links to the cytoskeleton through nuclear envelope proteins.
Isolated telomeric fragments do not behave as though they contain single-stranded DNA; instead, they show aberrant electrophoretic mobility and other properties.
Guanine bases have an unusual capacity to associate with one another. The single-stranded G-rich tail of the telomere can form G-quadruplex (also called G4 DNA or G quartets) of G residues. Each quartet contains four guanines that hydrogen bond with one another to form a planar structure. Each guanine comes from the corresponding position in a successive TTAGGG repeating unit.
FIGURE 1 shows an organization based on a crystal structure. The quartet that is illustrated represents an association between the first guanine in each repeating unit. It is stacked on top of another quartet that has the same organization, but is formed from the second guanine in each repeating unit. A series of quartets could be stacked like this in a helical manner. Although the formation of this structure attests to the unusual properties of the G-rich sequence in vitro, it does not demonstrate whether the quartet forms in vivo, for which there is only limited evidence to date.
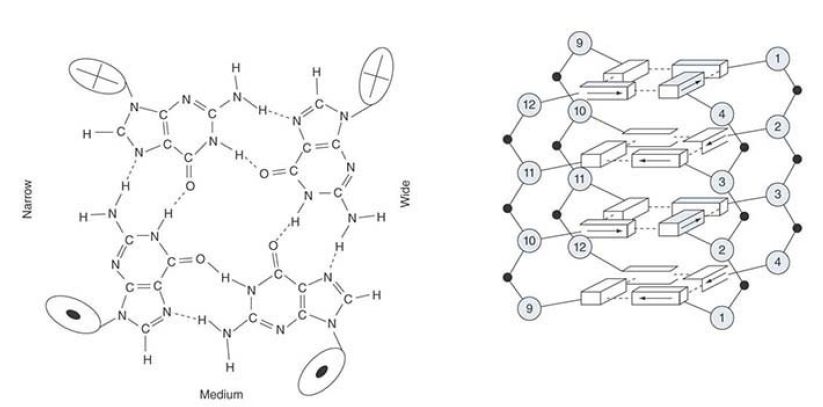
FIGURE 1. The crystal structure of a short repeating sequence from the human telomere forms three stacked G quartets. The top quartet contains the first G from each repeating unit. This is stacked above quartets that contain the second G (G3, G9, G15,G21) and the third G (G4, G10, G16, G22).
What feature of the telomere is responsible for the stability of the chromosome end? The schematic in FIGURE 2 shows that a loop of DNA forms at the telomere. The absence of any free end might be the crucial feature that stabilizes the end of the chromosome. The average length of the loop in animal cells is 5 to 10 kb. The loop is formed when the 3′ single-stranded end of the telomere (TTAGGG)n displaces the same sequence in an upstream region of the telomere. This converts the duplex region into a structure called a t-loop, where a series of TTAGGG repeats are displaced to form a single-stranded region, and the tail of the telomere is paired with the homologous strand.
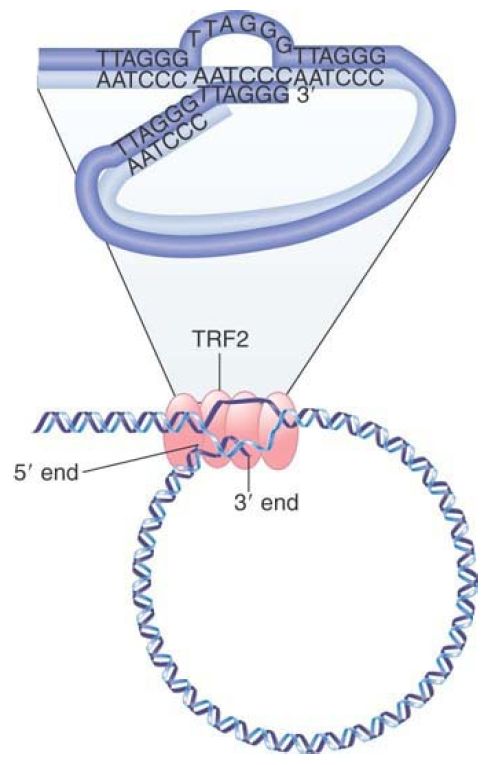
FIGURE 2. A loop forms at the end of chromosomal DNA. The 3′ single-stranded end of the telomere (TTAGGG)n displaces the homologous repeats from duplex DNA to form a t-loop. The reaction is catalyzed by TRF2.
© Dr. Gopal Murti/Science Source.
The reaction is catalyzed by the telomere-binding protein TRF2, which together with other proteins forms a complex that stabilizes the chromosome ends. Its importance in protecting the ends is indicated by the fact that the deletion of TRF2 causes chromosome rearrangements to occur.
In mammals, six telomeric proteins (TRF1, TRF2, Rap1, TIN2,TPP1, and POT1) primarily comprise a complex called shelterin, depicted in FIGURE 3 Shelterin functions to protect telomeres from DNA damage repair pathways and to regulate telomere length control by telomerase (discussed in the next section). Increasing roles for telomeres in aging, cancer, and cell differentiation reveal that telomeres are more than static caps at the ends of linear chromosomes.
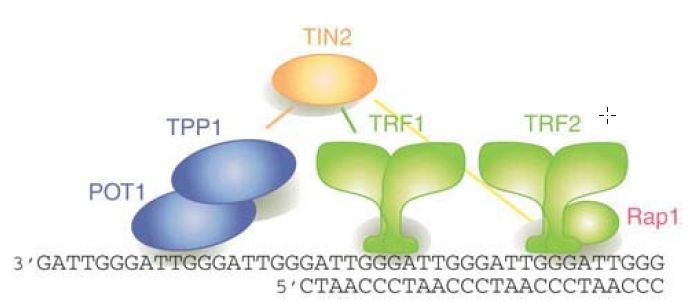
FIGURE 3 A schematic of how shelterin might be positioned on telomeric DNA, highlighting the duplex telomeric DNA interactions of TRF1 and TRF2 and the binding of POT1 to the single-stranded TTAGGG repeats. Although one of the shelterin complexes may have the depicted structure, telomeres contain numerous copies of the complex bound along the double-stranded TTAGGG repeat array. It is not known whether all (or even most) shelterins are present in six-protein complexes. Nucleosomes are omitted for simplicity.
Reprinted with permission from the Annual Review of Genetics, Volume 42 © 2008 by Annual Reviews www.annualreviews.org. Courtesy of Titia de Lange, The Rockefeller University.
Besides their role in capping the ends of linear chromosomes, telomeres also have an ancient and conserved function in meiosis, whereby they cluster on the nuclear envelope just prior to
homologous chromosome synapsis. This clustering defines the “bouquet” stage of meiosis, as shown in FIGURE 4, and represents a once-in-a-life-cycle configuration. The telomere clustering involves motility forces that act across the nuclear envelope via microtubules, actin, or other filamentous systems.
Genetic disruption of meiotic telomere clustering results in chromosome recombination and segregation defects, including the production of aneuploid daughter cells or sterility. Interestingly, fruit flies, which lack canonical telomerase-based telomeres, do not exhibit meiotic telomere clustering, but have evolved other mechanisms to ensure homologous chromosome pairing.
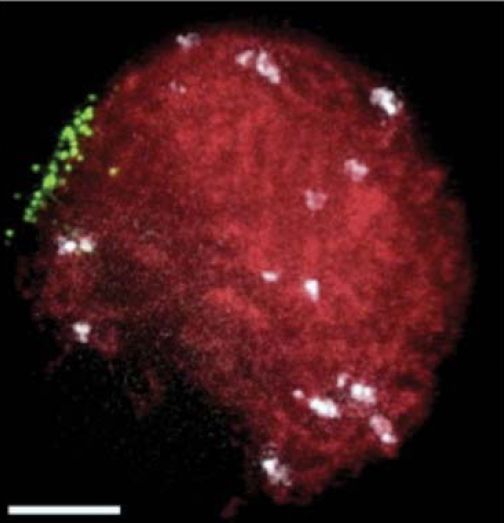
FIGURE 4. The meiotic telomere cluster is visualized by telomere FISH. Microscopic image of a maize nucleus fixed at meiotic prophase (zygotene stage), subjected to telomere (green) and centromere (white) FISH, and counter-stained for total DNA with DAPI (red). This pseudocolored image is a two-dimensional projection of a three-dimensional, multi-color image dataset.
Photo courtesy of S. P. Murphy and H. W. Bass, Florida State University.

|
|
|
|
مقاومة الأنسولين.. أعراض خفية ومضاعفات خطيرة
|
|
|
|
|
|
|
أمل جديد في علاج ألزهايمر.. اكتشاف إنزيم جديد يساهم في التدهور المعرفي ؟
|
|
|
|
|
|
|
العتبة العباسية المقدسة تقيم ندوة علمية عن روايات كتاب نهج البلاغة
|
|
|